Error Metrics for Multi-class Problems in R: Beyond Accuracy and Kappa
The caret package for R provides a variety of error metrics for regression models and 2-class classification models, but only calculates Accuracy and Kappa for multi-class models. Therefore, I wrote the following function to allow caret::train to calculate a wide variety of error metrics for multi-class problems:
#Multi-Class Summary Function
#Based on caret::twoClassSummary
require(compiler)
require(pROC)
multiClassSummary <- cmpfun(function (data, lev = NULL, model = NULL){
#Load Libraries
require(Metrics)
require(caret)
#Check data
if (!all(levels(data[, "pred"]) == levels(data[, "obs"])))
stop("levels of observed and predicted data do not match")
#Calculate custom one-vs-all stats for each class
prob_stats <- lapply(levels(data[, "pred"]), function(class){
#Grab one-vs-all data for the class
pred <- ifelse(data[, "pred"] == class, 1, 0)
obs <- ifelse(data[, "obs"] == class, 1, 0)
prob <- data[,class]
#Calculate one-vs-all AUC and logLoss and return
cap_prob <- pmin(pmax(prob, .000001), .999999)
prob_stats <- c(auc(obs, prob), logLoss(obs, cap_prob))
names(prob_stats) <- c('ROC', 'logLoss')
return(prob_stats)
})
prob_stats <- do.call(rbind, prob_stats)
rownames(prob_stats) <- paste('Class:', levels(data[, "pred"]))
#Calculate confusion matrix-based statistics
CM <- confusionMatrix(data[, "pred"], data[, "obs"])
#Aggregate and average class-wise stats
#Todo: add weights
class_stats <- cbind(CM$byClass, prob_stats)
class_stats <- colMeans(class_stats)
#Aggregate overall stats
overall_stats <- c(CM$overall)
#Combine overall with class-wise stats and remove some stats we don't want
stats <- c(overall_stats, class_stats)
stats <- stats[! names(stats) %in% c('AccuracyNull',
'Prevalence', 'Detection Prevalence')]
#Clean names and return
names(stats) <- gsub('[[:blank:]]+', '_', names(stats))
return(stats)
})
This function was prompted by a question on cross-validated, asking what the optimal value of k is for a knn model fit to the iris dataset. I wanted to look at statistics besides accuracy and kappa, so I wrote a wrapper function for caret::confusionMatrix and auc and logLoss from the Metric packages. Use the following code to fit a knn model to the iris dataset:
library(caret)
set.seed(19556)
model <- train(
Species~.,
data=iris,
method='knn',
tuneGrid=expand.grid(.k=1:30),
metric='Accuracy',
trControl=trainControl(
method='repeatedcv',
number=10,
repeats=15,
classProbs=TRUE,
summaryFunction=multiClassSummary))
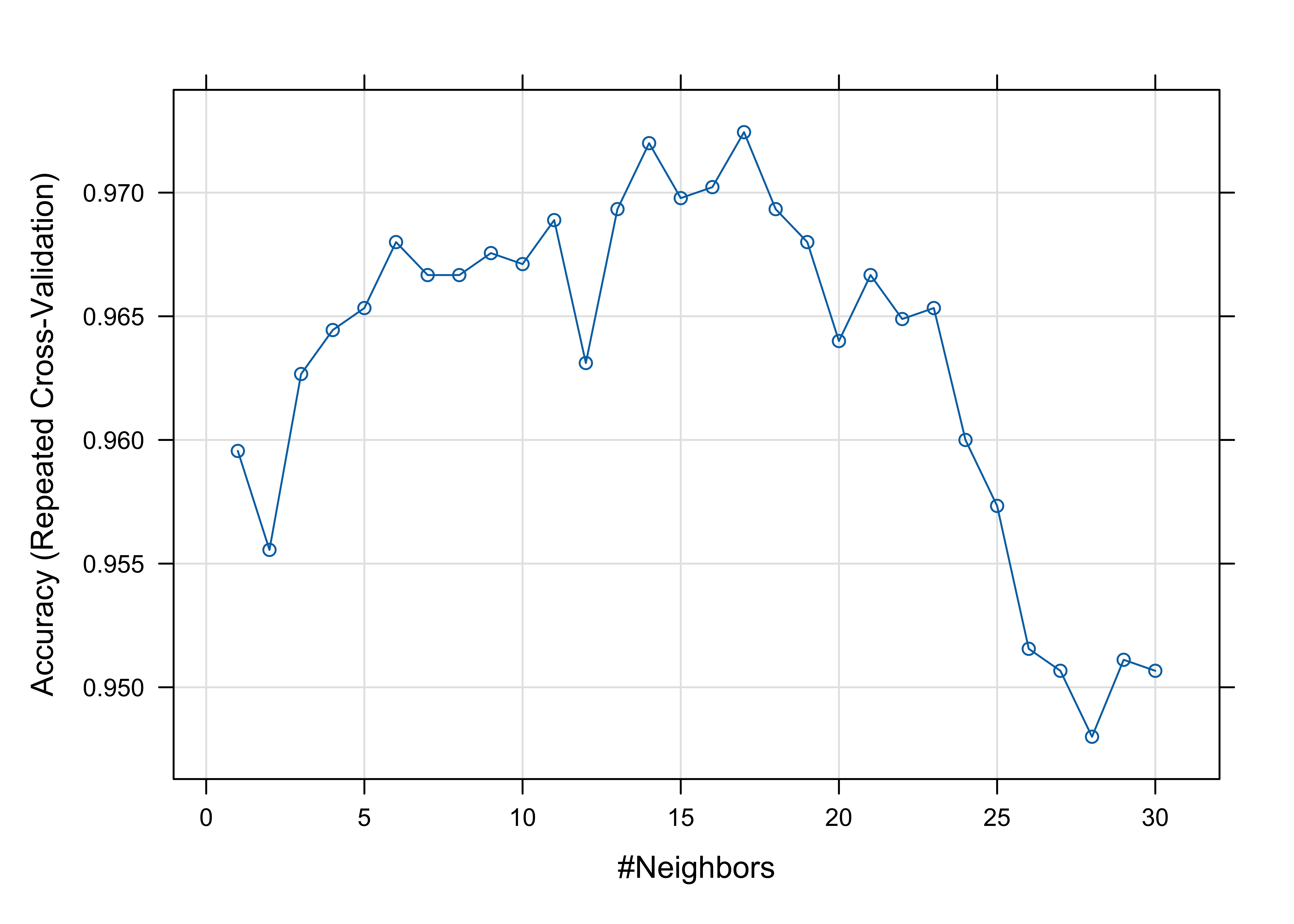
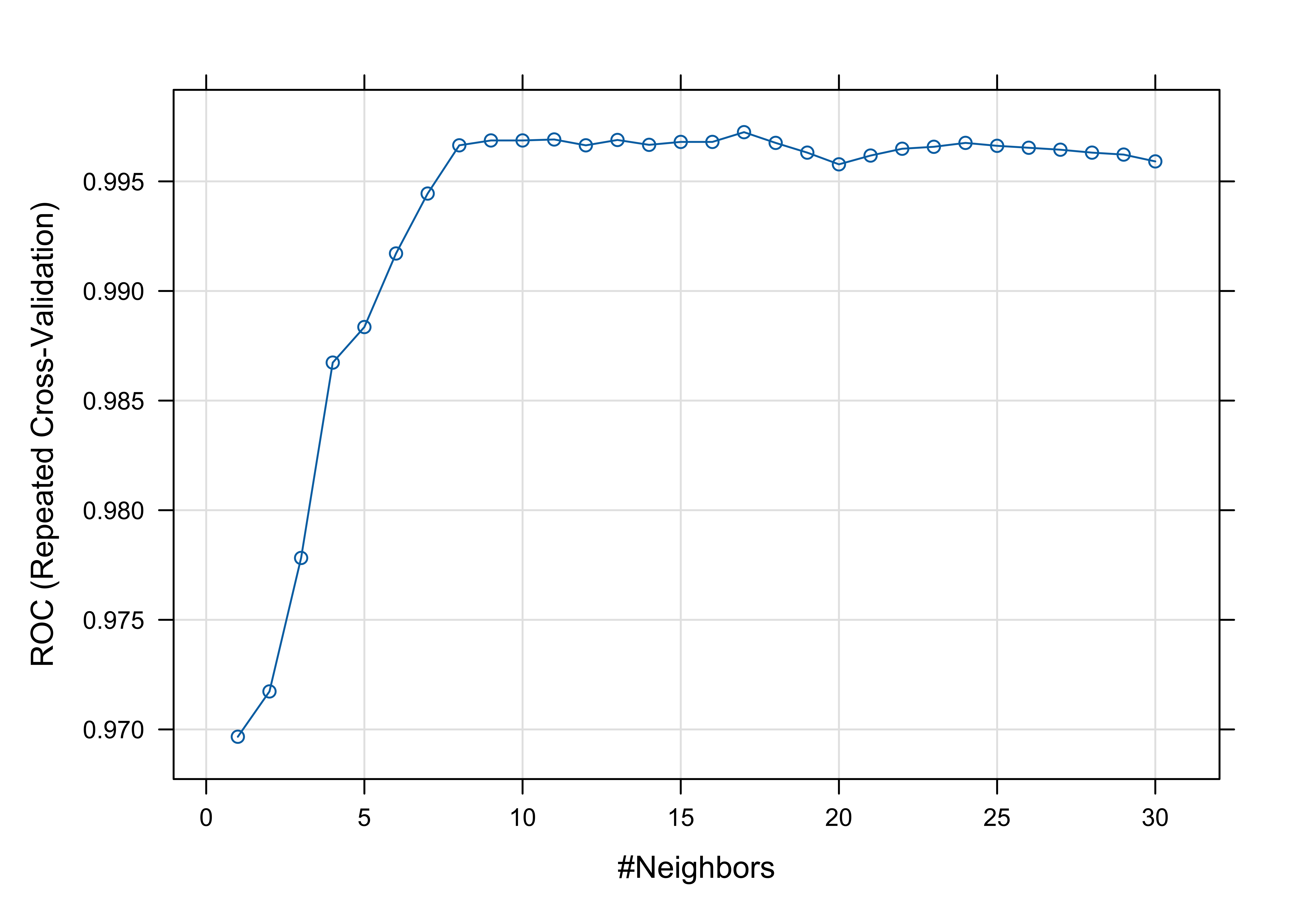
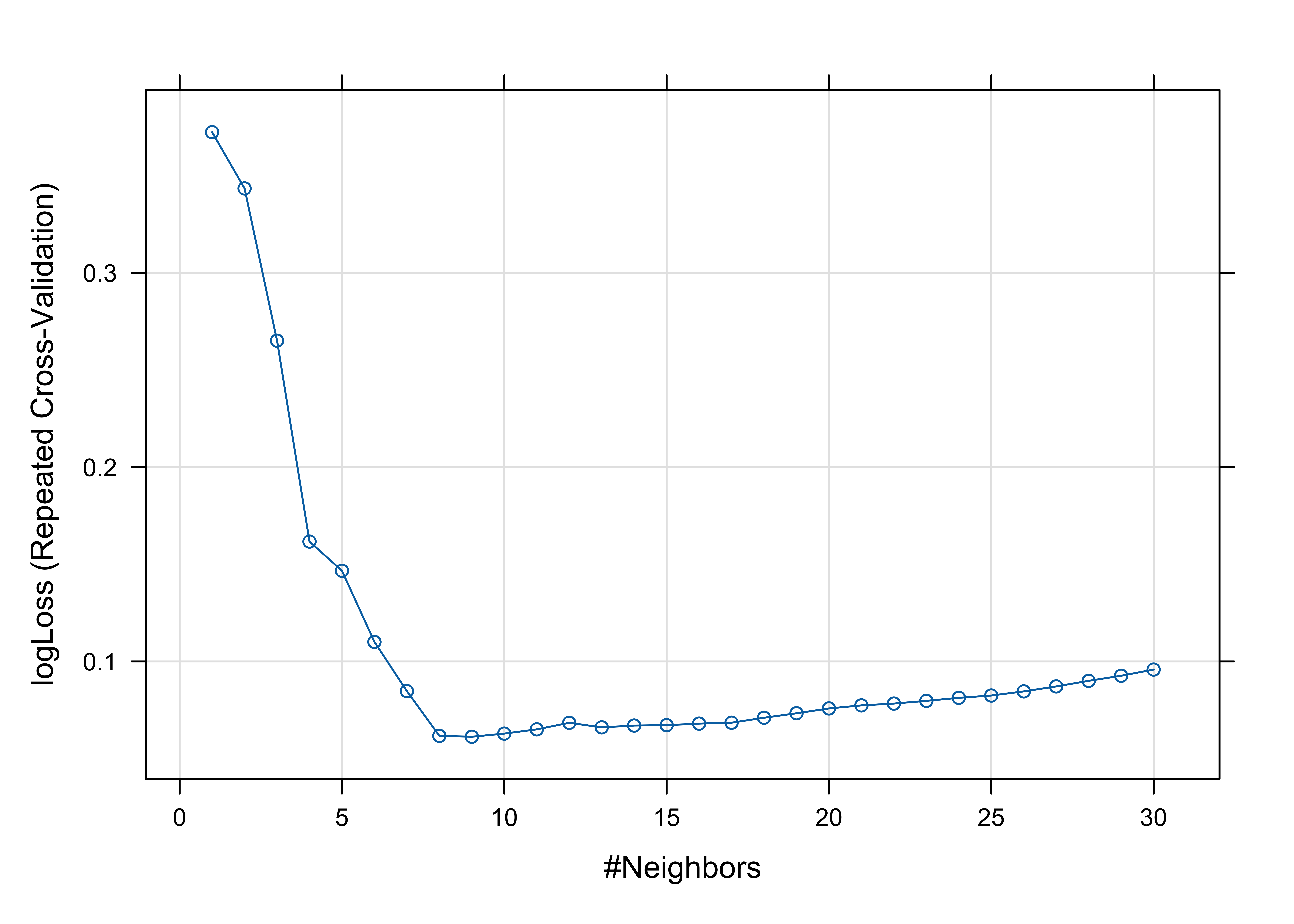
This demonstrates that, depending on what metric you use, you will end up with a different model. For example, Accuracy seems to peak around 17, while AUC and logLoss seem to peak around 6:
# All possible metrics:
# c('Accuracy', 'Kappa', 'AccuracyLower', 'AccuracyUpper', 'AccuracyPValue',
# 'Sensitivity', 'Specificity', 'Pos_Pred_Value',
# 'Neg_Pred_Value', 'Detection_Rate', 'ROC', 'logLoss')
print(plot(model, metric='Accuracy'))
print(plot(model, metric='ROC'))
print(plot(model, metric='logLoss'))
You can also increase the number of cross-validation repeats, or use a different method of re-sampling, such as bootstrap re-sampling.