caretEnsemble 4.0
caretEnsemble is a package for creating stacked ensembles of models in R. I’ve been working on this package for over a decade, and I’m proud that it’s still useful even after all these years!
I first wrote this package when I was active on Kaggle, as model stacks are a common technique for winning Kaggle competitions. I loved using the caret package to cross-validate models in R, and caretEnsemble was born out of all my custom R scripts for stacking caret models. I wrote the first draft of caretEnsemble when I was on an Amtrak train from New York to Boston in 2013, and I’ve been maintaining it ever since.
caretEnsemble 4.0, the first update in years, introduces several new features. The highlight is the reintroduction of the greedy ensembling algorithm. This algorithm, present in version 0.0 but removed from the 1.0 CRAN submission due to complexity, has been reimplemented in a clean, modular way. I’m proud of the new implementation: it will be much more maintainable in the future.
This greedy selection algorithm was first described in Ensemble Selection from Libraries of Models and it has some nice properties: 1. The weights of the component models are always positive (negative weights can lead to odd results on out-of-sample data). 2. If it is hard to find a good ensemble, the greedy optimizer will simply select the best single model.
At the end of the day, it’s a simple weighted average of the input models, which turns out to be a robust way to combine models. But if you like complexity, you can also use caretEnsemble to do more complex stacks, using any caret model for the ensemble model. Many winning Kaggle solutions used XGboost to ensemble a variety of input models.
Version 4.0 adds a lot of new features, including greedy stacks, multiclass support, support for mixed-type ensembles, and transfer learning.
Greedy Ensemble demo
First, you will likely need to install the package from CRAN:
install.packages("caretEnsemble", repos = "https://cloud.r-project.org/")
For this demo, I’m going to use the “AmesHousing” and the ranger package, so you should install those too.
install.packages(c("AmesHousing", "Xgboost"), repos = "https://cloud.r-project.org/")
Take a quick look at the data:
set.seed(42)
dat <- AmesHousing::make_ames()
head(dat)
#> # A tibble: 6 × 81
#> MS_SubClass MS_Zoning Lot_Frontage Lot_Area Street Alley Lot_Shape Land_Contour Utilities Lot_Config Land_Slope
#> <fct> <fct> <dbl> <int> <fct> <fct> <fct> <fct> <fct> <fct> <fct>
#> 1 One_Story_1946_an… Resident… 141 31770 Pave No_A… Slightly… Lvl AllPub Corner Gtl
#> 2 One_Story_1946_an… Resident… 80 11622 Pave No_A… Regular Lvl AllPub Inside Gtl
#> 3 One_Story_1946_an… Resident… 81 14267 Pave No_A… Slightly… Lvl AllPub Corner Gtl
#> 4 One_Story_1946_an… Resident… 93 11160 Pave No_A… Regular Lvl AllPub Corner Gtl
#> 5 Two_Story_1946_an… Resident… 74 13830 Pave No_A… Slightly… Lvl AllPub Inside Gtl
#> 6 Two_Story_1946_an… Resident… 78 9978 Pave No_A… Slightly… Lvl AllPub Inside Gtl
#> # ℹ 70 more variables: Neighborhood <fct>, Condition_1 <fct>, Condition_2 <fct>, Bldg_Type <fct>, House_Style <fct>,
#> # Overall_Qual <fct>, Overall_Cond <fct>, Year_Built <int>, Year_Remod_Add <int>, Roof_Style <fct>, Roof_Matl <fct>,
#> # Exterior_1st <fct>, Exterior_2nd <fct>, Mas_Vnr_Type <fct>, Mas_Vnr_Area <dbl>, Exter_Qual <fct>, Exter_Cond <fct>,
#> # Foundation <fct>, Bsmt_Qual <fct>, Bsmt_Cond <fct>, Bsmt_Exposure <fct>, BsmtFin_Type_1 <fct>, BsmtFin_SF_1 <dbl>,
#> # BsmtFin_Type_2 <fct>, BsmtFin_SF_2 <dbl>, Bsmt_Unf_SF <dbl>, Total_Bsmt_SF <dbl>, Heating <fct>, Heating_QC <fct>,
#> # Central_Air <fct>, Electrical <fct>, First_Flr_SF <int>, Second_Flr_SF <int>, Low_Qual_Fin_SF <int>,
#> # Gr_Liv_Area <int>, Bsmt_Full_Bath <dbl>, Bsmt_Half_Bath <dbl>, Full_Bath <int>, Half_Bath <int>, …
To make things a little easier for this demo, we’ll model the log of the sale price, and also center and scale it:
dat$Sale_Price <- log(dat$Sale_Price)
dat$Sale_Price <- scale(dat$Sale_Price, center=TRUE, scale=TRUE)
Now let’s split it into a training set and a test set:
set.seed(780L)
train_index <- caret::createDataPartition(dat$Sale_Price, p = 0.8, list = FALSE)
train_data <- dat[train_index, ]
test_data <- dat[-train_index, ]
Now that we have a train/test split, we can use caretList to train a set of base models. We use the ranger package to fit a random forest, which requires no processing and works well out-of-the-box. We also fit a simple linear regression model with some preprocessing steps (removing zero-variance predictors and applying PCA. Zero variance predictors and correlated predictors can both lead to issues with linear models).
These models take a few minutes to train, so grab a cup of coffee while you wait!
set.seed(506L)
model_list <- caretEnsemble::caretList(
Sale_Price ~ .,
data = train_data,
methodList = "ranger",
tuneList = list(caretEnsemble::caretModelSpec(method = "lm", preProcess = c("zv", "pca")))
)
summary(model_list)
#> The following models were ensembled: lm, ranger
#>
#> Model accuracy:
#> model_name metric value sd
#> <char> <char> <num> <num>
#> 1: lm RMSE 0.3624948 0.04756348
#> 2: ranger RMSE 0.3428007 0.05774693
When looking at these results, remember that lower is better for RMSE.
Stacking these models is easy:
set.seed(961L)
greedy_stack <- caretEnsemble::caretEnsemble(
model_list,
tuneLength = 1
)
summary(greedy_stack)
#> The following models were ensembled: lm, ranger
#>
#> Model Importance:
#> lm ranger
#> 0.4476 0.5524
#>
#> Model accuracy:
#> model_name metric value sd
#> <char> <char> <num> <num>
#> 1: ensemble RMSE 0.3199006 0.04130564
#> 2: lm RMSE 0.3624948 0.04756348
#> 3: ranger RMSE 0.3428007 0.05774693
You can directly view the weights for the weighted average:
print(round(greedy_stack$ens_model$finalModel$model_weights, 3))
#> [,1]
#> lm 0.43
#> ranger 0.57
If you want a non-linear stack, you can use any model from the caret package. E.g. here’s how you would use a glm:
set.seed(961L)
glm_stack <- caretEnsemble::caretStack(
model_list,
method = "glm",
tuneLength = 1
)
summary(glm_stack)
#> The following models were ensembled: lm, ranger
#>
#> Model Importance:
#> lm ranger
#> 0.4112 0.5888
#>
#> Model accuracy:
#> model_name metric value sd
#> <char> <char> <num> <num>
#> 1: ensemble RMSE 0.3174498 0.04087683
#> 2: lm RMSE 0.3624948 0.04756348
#> 3: ranger RMSE 0.3428007 0.05774693
The glm is less constrained in the weights that it learns:
print(round(coef(glm_stack$ens_model$finalModel), 3))
#> (Intercept) lm ranger
#> 0.001 0.413 0.635
We can predict on new data with the stack and both ensembles:
preds <- data.table::data.table(predict(model_list, newdata = test_data))
preds[,greedy := predict(greedy_stack, newdata = test_data)]
preds[,glm := predict(glm_stack, newdata = test_data)]
And then calculate RMSE:
rmse <- sapply(preds, function(x) sqrt(mean((x - test_data$Sale_Price)^2)))
print(round(sort(rmse), 3))
#> glm greedy lm ranger
#> 0.311 0.313 0.325 0.342
Both ensembles do better on the test data than the base models.
Plots
caretEnsemble includes some useful plotting utilities. Use the base plot function to show the error of each model in the stack, as well as the overall error for the stack itself:
plot(glm_stack)
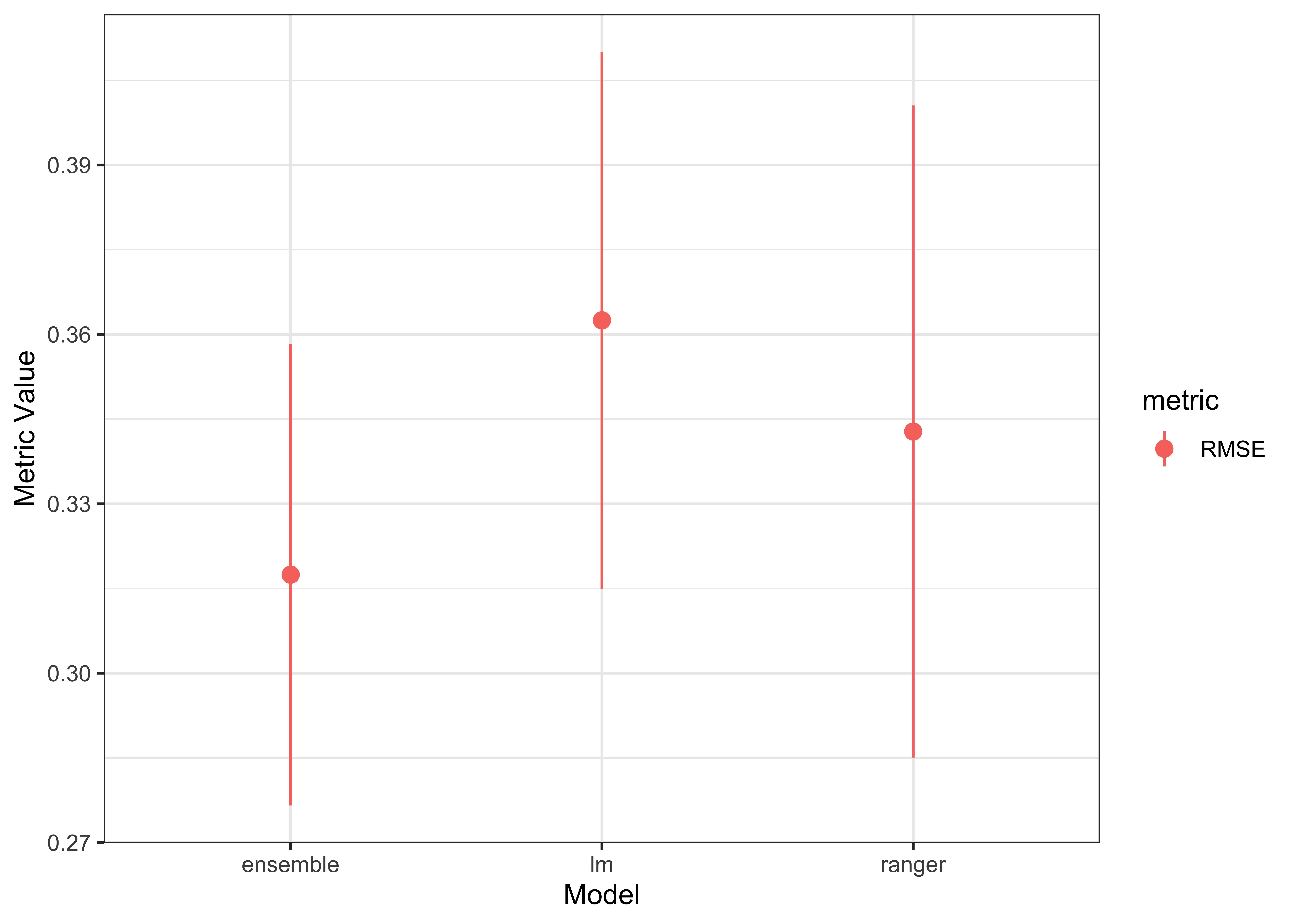
Note that while the linear model is generally worse and higher variance than the ranger model, including it in the stack both improves the ensemble and lowers the ensemble’s variance.
You can also use autoplot from ggplot2 to plot several diagnostics simultaneously:
ggplot2::autoplot(glm_stack, training_data=train_data, xvars=c("Year_Built", "Gr_Liv_Area"))
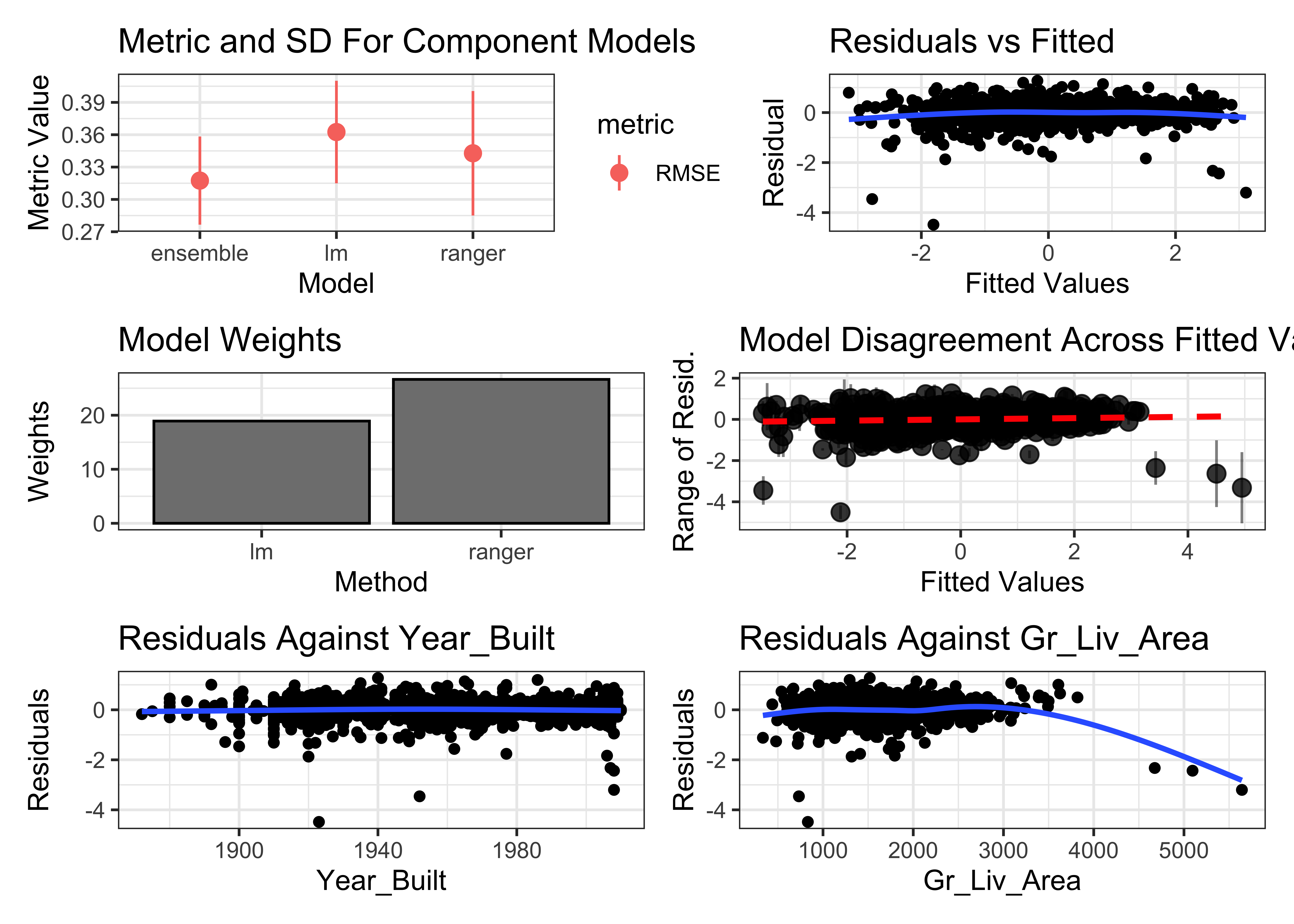
The 5 outliers in the training data with high residuals would be a good place to start investigating this model’s performance.
Further reading
A Brief Introduction to caretEnsemble walks through the basics of model stacking with caretEnsemble, while Version 4.0 New Features is an overview of all the new features in version 4.0.
You can also checkout the source code on GitHub. Make a pull request if you spot any bugs!